Dec 07, 2023
Penn State researchers announce new comprehensive -Omics database
A group of Penn State scientists have announced kmerDB, an extensive catalogue of sequence information representing all domains of life.
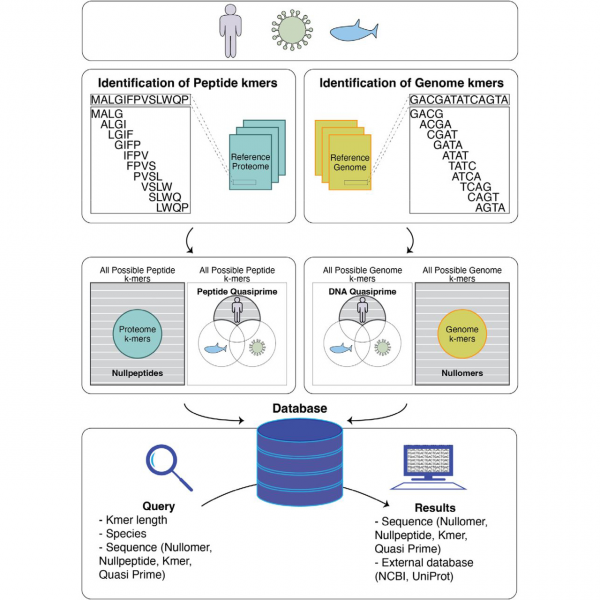
UNIVERSITY PARK, Pa.—In a new bioRxiv preprint article, a group of Penn State scientists have announced kmerDB, an extensive catalogue of sequence information representing all domains of life. The preprint, “kmerDB: A Database Encompassing the Set of Genomic and Proteomic Sequence Information for Each Species,” led by Ioannis Mouratidis, a research scientist in the Georgakopoulos-Soares Lab, was released on November 16, 2023.
Advances in sequencing and computational technologies over the past two decades have yielded genomes and proteomes for thousands of species. These DNA and protein sequences can be broken into simpler components called kmers, defined as subsequences of fixed lengths, for common applications such as metagenomic characterization and assembly quality control. However, increasingly diverse applications—such as disease diagnostics—will depend on a comparative catalogue to identify the shortest kmers that are present in a single species (quasi-primes), absent from all species (primes), or absent from a sample (nullomers).
KmerDB addresses this gap by providing a curated listing of kmer, nullomer, nullpeptide, quasi-prime, and prime sequences for 45,785 species and 22,386 proteomes via a public web interface. Users can easily browse and query the database using species name, NCBI ID, Uniprot ID, or conduct advanced searches of various taxonomic levels and kmer counts. The database also contains useful metrics such as the polarity and charge of protein kmers and the nucleotide composition for DNA kmers.
The authors highlight several research applications that kmerDB may facilitate such as pathogen detection, cancer detection, sample barcoding (using prime sequences) and forensic applications. Additionally, kmerDB can prove useful for evolutionary genomics research and biotechnology applications such as developing CRISPR target sites.
“By providing a pre-computed kmer database that would take months of computational time and hundreds of computer processors to calculate, we hope to spur further research into the fields of kmers, nullomers, and quasi-primes and their many applications,” said Ilias Georgakopoulos-Soares.
“This work is the result of imagining a new genetic resource for life’s diversity - a comprehensive database of DNA and protein fingerprints that enable biologists worldwide to track disease variants, pathogens, microbiome communities, and evolution. Biology will be well served by the impact kmerDB brings to both basic and applied research”, said Seth Bordenstein, director of the One Health Microbiome Center, Huck Endowed Chair of the Microbiome Sciences, and professor of Biology and Entomology.
Other Penn State authors on the paper include Ilias Georgakopoulos-Soares, Maxwell A. Konnaris, Nikol Chantzi, Jasna Kovac and Austin Montgomery. Additional authors include Fotis A. Baltoumas and Georgios A. Pavlopoulos, National and Kapodistrian University of Athens; Candace S.Y. Chan, University of California San Francisco; George C. Georgakopoulos, National Technical University of Athens; Dionysios Chartoumpekis, Lausanne University.